New insights into molecular machines through biophysical methods
Researchers at the University of Regensburg have gained groundbreaking insights into the movements of the RNA-exosome complex, a molecular machine responsible for the degradation of RNA in cells. By combining cryo-electron microscopy, nuclear magnetic resonance spectroscopy (NMR) and molecular dynamics simulations (MD), the interdisciplinary team succeeded in investigating the structure and dynamics of this protein complex in detail. The results, published in the journal Nature Communications, mark a methodological advance in the study of large molecular machines.
Proteins are essential for all life processes and often form complexes that perform a variety of tasks as molecular machines, such as the assembly and degradation of biomolecules or transport within cells. The RNA exosome, consisting of ten proteins, plays a central role in the degradation of RNA, an indispensable process in every cell. While the static structure of such complexes can be well studied by techniques such as cryo-electron microscopy, capturing their movements has been a challenge so far. The team from the Regensburg Center for Biochemistry (RCB) and the Regensburg Center for Ultrafast Nanoscopy (RUN) has now made the dynamic processes of the exosome visible for the first time.
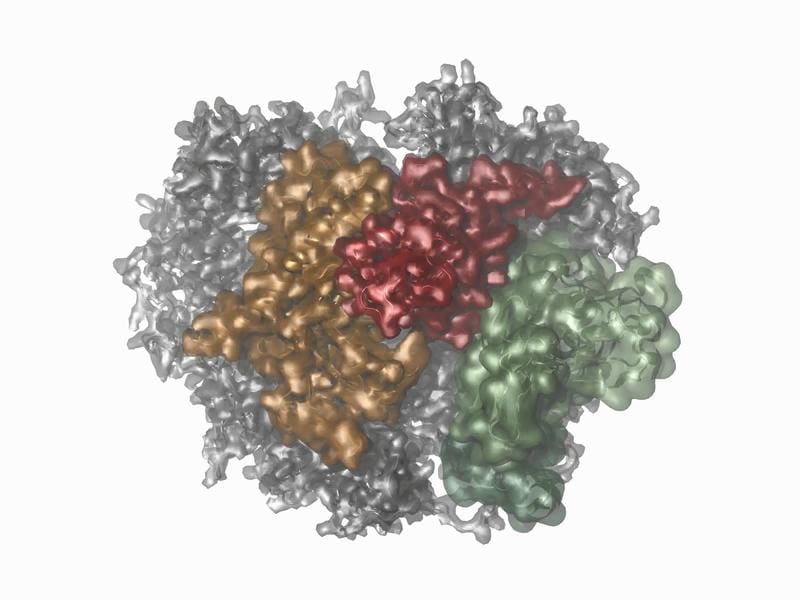
Using NMR spectroscopy, the researchers investigated how the structure of the proteins changes during their function. This method, which is normally used for smaller proteins, could also be applied to the large exosome complex through innovative approaches. Molecular dynamics simulations complemented the NMR data by providing vivid models of protein motion. This allowed the scientists to identify regions of the complex that were previously invisible to other methods and analyze their movements ŌĆō from extremely fast oscillations in the billion-second range to slower motions that occur about 30 times per second.
Particularly revealing was the discovery that some protein regions move in a rhythm that correlates with the rate of RNA degradation. This suggests that the dynamics of the proteins are crucial for their function. The study thus not only provides new insights into the RNA degradation process, but also establishes a method to visualize the movements of large molecular machines. This approach could revolutionize research on other protein complexes and create the basis for a deeper understanding of dynamic processes in cells.
Original Paper:
4D structural biologyŌĆōquantitative dynamics in the eukaryotic RNA exosome complex | Nature Communications
Editor: X-Press Journalistenb├╝ro GbR
Gender Notice. The personal designations used in this text always refer equally to female, male and diverse persons. Double/triple naming and gendered designations are used for better readability. ected.




