Zoonoses: CRISPR-Cas systems revolutionize pathogen detection
The rapid spread of animal diseases and the evolution of pathogens pose a significant threat to animal husbandry. Traditional detection methods such as PCR require expensive equipment and specialized personnel, making them unsuitable for use on farms or in rural areas. In recent years, the CRISPR-Cas system, originally developed for gene editing, has transformed into an innovative detection tool. In particular, the class 2 proteins Cas9, Cas12 and Cas13 enable fast and sensitive detection on site due to their precise recognition of nucleic acid sequences. Combined with isothermal amplification technology, they offer efficient solutions for pathogen detection in animals.
A review by a team from the Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry at Tianjin Agricultural University highlights class 2 CRISPR-Cas systems as a major breakthrough. These systems work with a single Cas protein, which allows for simple structure and easy modification. In combination with isothermal amplification, detection takes place in a short time. In the Cas12 system, the discovery of the target DNA activates collateral fission, which severs fluorescent probes or labeled molecules on test strips and indicates results through signal changes. The work was published in the journal Frontiers of Agricultural Science and Engineering.
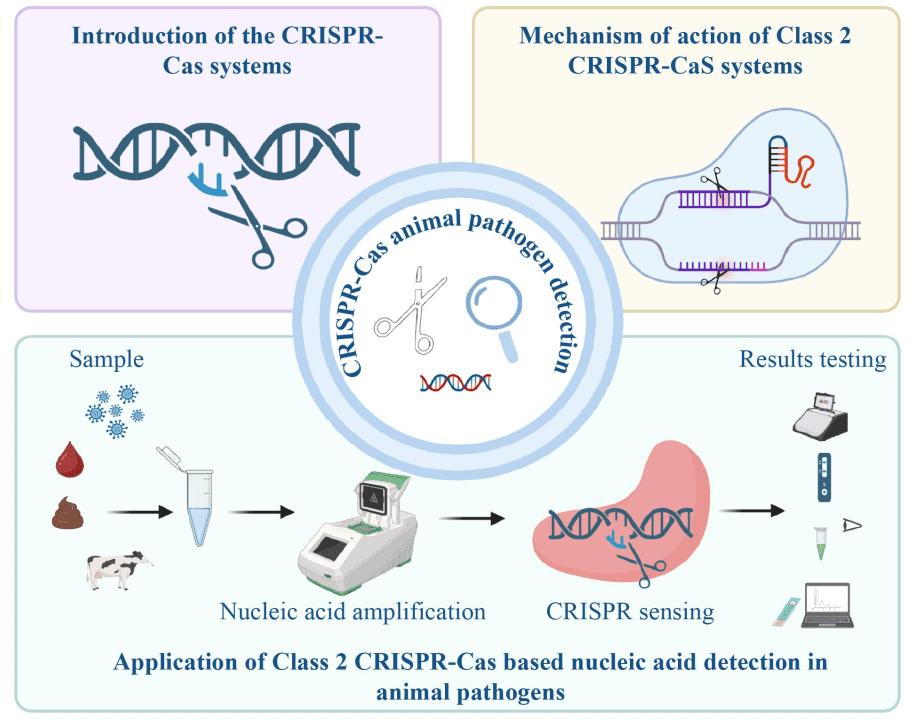
For RNA viruses such as the bird flu virus or the duck tembusu virus, the Cas13 system performs outstandingly. The SHERLOCK platform combines recombinase polymerase amplification with transcription to amplify target RNA, followed by precise detection by Cas13, which triggers fluorescent signals. Studies show that the detection of the H5 subtype of the bird flu virus with Cas13 and RT-RAA takes only 40 minutes, with a sensitivity of 0.1 virus copy per microliter. In combination with lateral flow strips, results can be read without equipment, which supports basic veterinarians in the field. Cas13 also enables the simultaneous detection of multiple pathogens, such as duck hepatitis A virus type 3 and new duck reovirus, which increases diagnostic efficiency.
The core advantages lie in its suitability for the field: While PCR takes several hours, most CRISPR-based methods are completed within an hour. They achieve a high sensitivity down to the single copy level, as in the case of Cas12 detection of the porcine reproductive and respiratory syndrome virus in less than 25 minutes with only one virus copy, which avoids false negatives. No expensive equipment is necessary; Operations are performed at simple heating or room temperature, and results are displayed visually, for example by fluorescence or color change on test strips. When African swine fever is detected, farmers receive results in half an hour with portable strips, allowing for rapid isolation of infected animals and preventing herd infections to minimize economic losses and contain epidemics.
However, the overview points to challenges: Some methods carry a risk of contamination due to nucleic acid amplification; Contaminants such as blood or feces in animal samples can interfere with detection. Future research directions include amplification-free CRISPR detections or optimized systems against sample contamination to ensure more reliable application in the field.
Overall, the Class 2 CRISPR-Cas system opens up new avenues for pathogen detection in animals. It promises to become a practical tool for epidemic prevention in animal husbandry, especially in resource-poor regions. Rapid diagnoses can protect the industry and promote healthy development. The technology overcomes the limitations of traditional processes by combining speed, sensitivity and simplicity. Researchers expect further adjustments to make the method more robust. Integration into routine procedures could improve global animal health and contain outbreaks at an early stage. The work highlights the transition from laboratory methods to field-based approaches, which is particularly relevant for developing countries.
Editor: X-Press Journalistenbû¥ro GbR
Gender Notice. The personal designations used in this text always refer equally to female, male and diverse persons. Double/triple naming and gendered designations are used for better readability. ected.




